Browse genomes and community contributed datasets. If you find a dataset missing or have a new dataset to contribute for a supported genome please contact us!
[+]S. purpuratus (Purple sea urchin) v.5.0
L. variegatus (Green sea urchin) v.3.0
A. planci (Crown-of-thorns) v.1.0
[+]L. pictus (Painted urchin) v.3.0
A. japonica (Feather star) v.1.0
BLAST Echinoderm nucleotide and protein databases.
Genome, RNA, Proteins, CDS
L. variegatus (Green sea urchin):
Genome RNA, Proteins, CDS
P. miniata (Bat star):
Genome , RNA, Proteins, CDS
A. planci (Crown-of-thorns):
Genome, RNA, Proteins, CDS
L. pictus (Painted urchin): Genome
A. rubens (Sugar star): Genome
A. japonica (Feather star): Genome
A. filiformis (Brittle star): Genome
P. flava (Yellow acorn worm): Genome
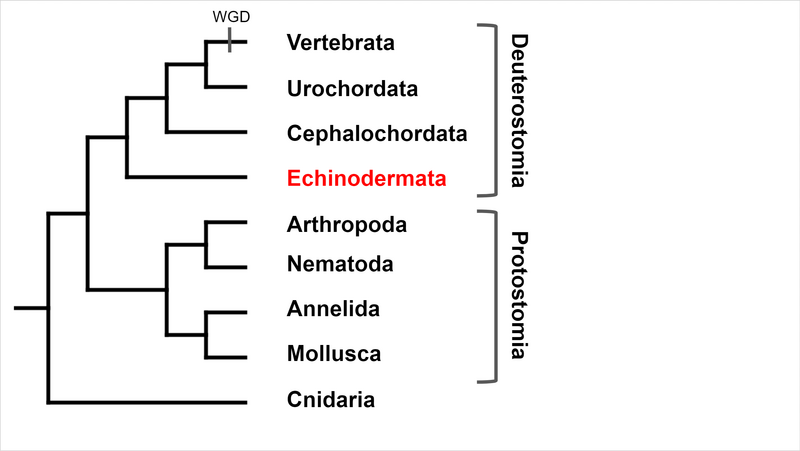
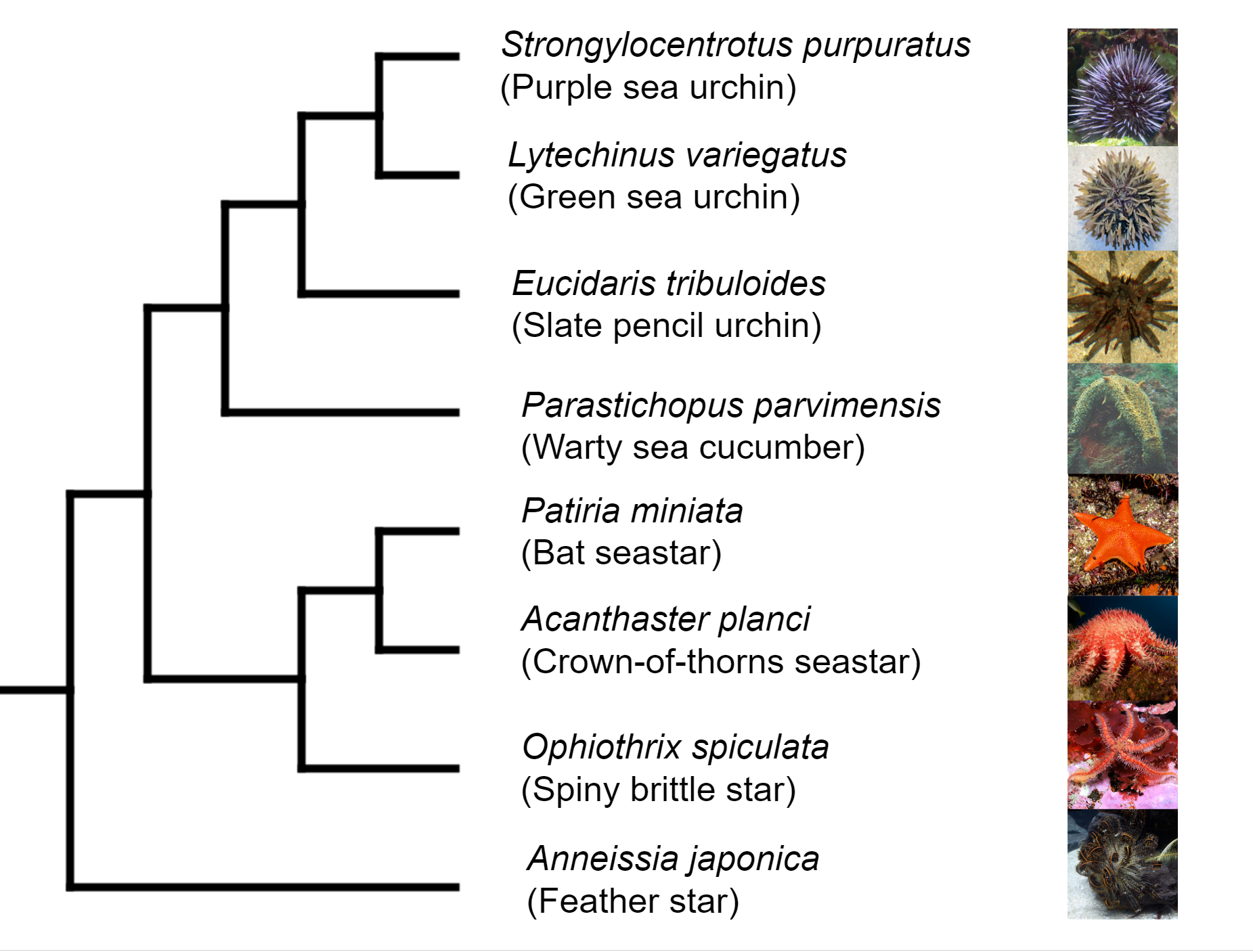 Deuterostomia (superphylum) includes Chordata, Echinodermata and
Hemichordata phylum, Echinodermata emerged in the Lower Cambrian
(> 540 million years ago, MYA) and split from Chordata 400-500
MYA, Echinodermata includes 7,000 extant and 13,000 extinct,
marine only species. Echinodermata has 5 Classes: the basal
branching Crinoidea (crinoids), and the 4 motile Eleutherozoa
Classes including Asterozoa, the Ophiuroidea (brittle stars) and
Asteroidea (starfish) and the Echinozoa, the Echinoidea (sea
urchins and sand dollars) and Holotheroidea (sea cucumber).
Deuterostomia (superphylum) includes Chordata, Echinodermata and
Hemichordata phylum, Echinodermata emerged in the Lower Cambrian
(> 540 million years ago, MYA) and split from Chordata 400-500
MYA, Echinodermata includes 7,000 extant and 13,000 extinct,
marine only species. Echinodermata has 5 Classes: the basal
branching Crinoidea (crinoids), and the 4 motile Eleutherozoa
Classes including Asterozoa, the Ophiuroidea (brittle stars) and
Asteroidea (starfish) and the Echinozoa, the Echinoidea (sea
urchins and sand dollars) and Holotheroidea (sea cucumber). Echinoderms are model organisms for studying embryo development and regeneration due to many unique features including:
- the ability to synchronize fertilization of millions of eggs
- transparent embryos and larvae
- varied development within and between a genus
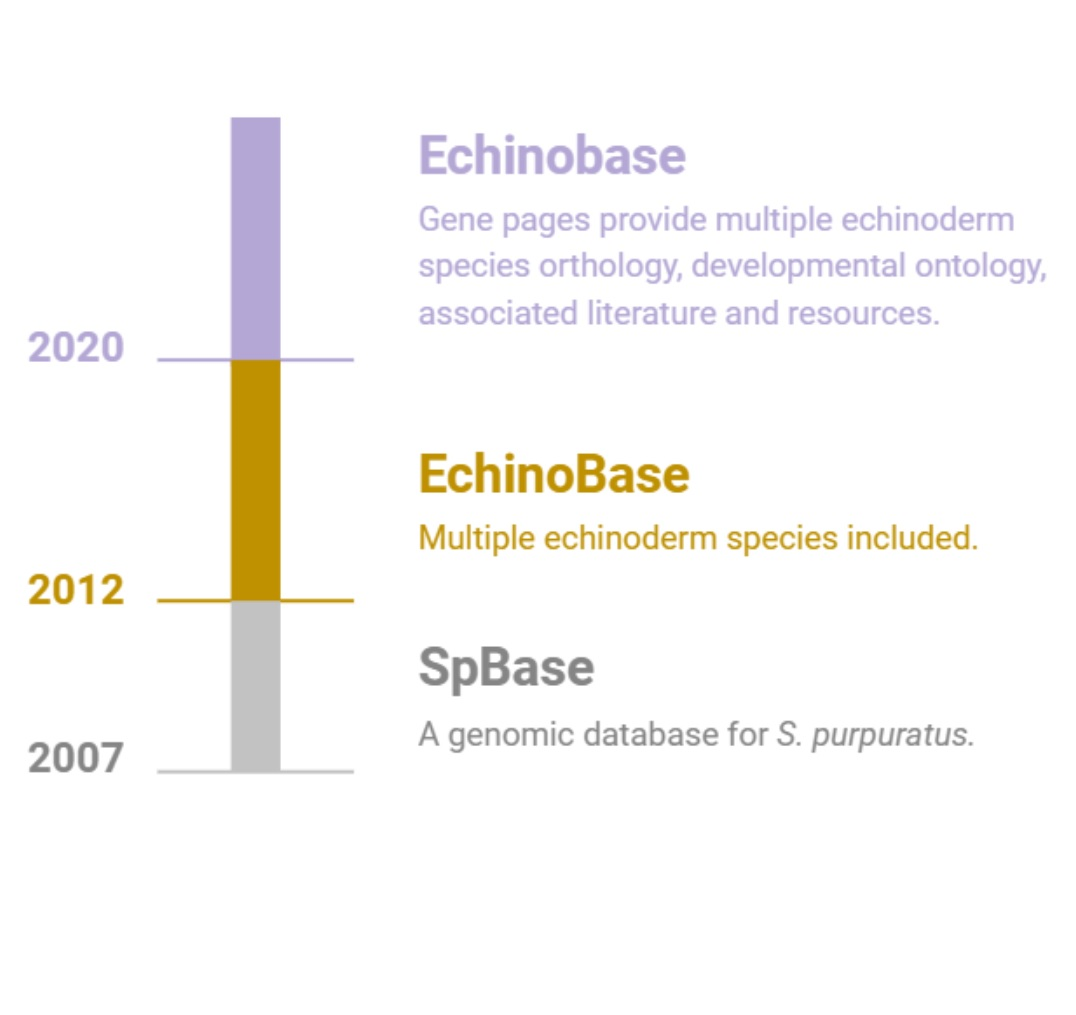 Echinobase supports the international research community by providing a centralized, integrated and easy to use web based
resource to access the diverse and rich, functional genomics data of echinoderm species.
Echinobase supports the international research community by providing a centralized, integrated and easy to use web based
resource to access the diverse and rich, functional genomics data of echinoderm species.Echinobase is organized around the GENEPAGE which displays information about genes, orthology, and links to research papers. The gene models of echinoderm species are associated with the genome sequence using the genome browser, JBrowse. Temporal developmental gene expression data will be displayed when available, and spatial cell type information will be associated using the Echinoderm Anatomy and Development Ontology (ECAO).
Echinobase provides a critical data sharing infrastructure for other NIH-funded projects. In addition to our primary goal of supporting echinoderm researchers, Echinobase enhances the availability and visibility of echinoderm data to the broader biomedical research community.
Species in Echinobase are divided into two categories:
fully supported species found on gene pages and partially supported species that only have support through the genome browser and BLAST processes. We are an expanding resource and both species sets will grow in upcoming months and years.

The substantially improved current release of the purple sea urchin genome assembly (Spur5.0) is sixth in the series.



Hemichordates are generally regarded as the sister group to echinoderms and occupy an important evolutionary position between vertebrates and invertebrates. This makes P. flava a valuable model organism for studying the genomic and evolutionary origins of chordate and deuterostome traits.

Stemming from its remarkable ability to regenerate arms in a matter of weeks it has become an emerging model for regeneration and stem cell biology in biomedical research. Research targets include:
- The cellular and molecular mechanisms underlying regeneration with links to neuroscience, stem cell biology and neuropeptide structure and function in the absence of a centralised nervous system.
- Comparative developmental events involving the evolution of biomineralization in the echinoderm clade.
- Support for phylogenomic analysis in an otherwise little studied group.
Development raw reads are available at NCBI accession PRJNA349786.


The current version, v3.0, of the assembly is fully supported at Echinobase.

The L. variegatus v3.0 genome is the first echinoderm assembly with chromosomal resolution.

Learn more about the genomics of the crown-of-thorns seastar.
Echinoderms are marine organisms that when harvested from the
wild, can live in aquariums and embryo and larval development has
been studied for over 100 years. Echinoderm eggs are fertilized
externally and embryos and larvae are transparent, enabling
detailed descriptions of the structure and timing of development.
The variation and comparison of developmental patterns of
related species is used to improve understanding of these vital
processes and their evolution in echinoderms and deuterostomes.
Echinoderms are models for studies of skeletogenesis and
regeneration.
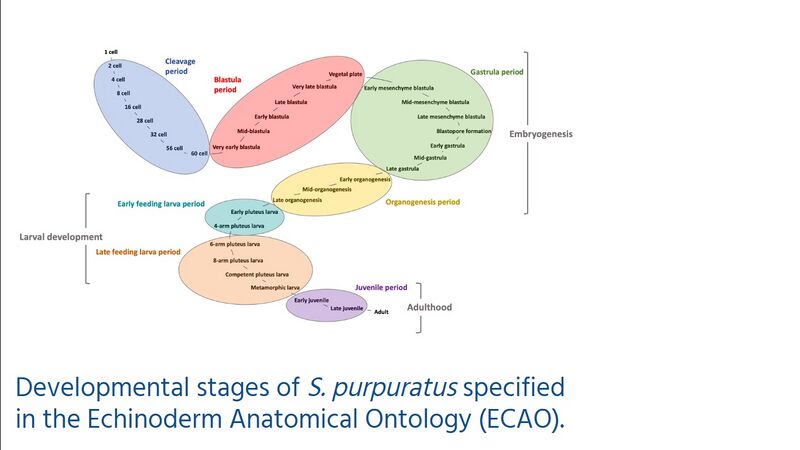
The Echinoderm
Anatomical Ontology (ECAO) provides standardized terms for
developmental stages for S.
purpuratus, and is being expanded to include additional
echinoderm species.

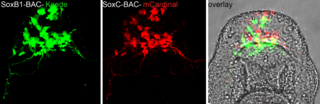
Echinobase has collected contact information for members of the community and is working to maintain a current, updated web interface, for accessing and querying genomes, transcriptomes, chromatin structure, regulation and expression of genes during development with documentation, and supporting information, an image database, and robust ontologies.
Community members are welcomed to contribute to EchinoWiki with protocols, reagents and data, and to submit new genomes for partial or full support.
Genomes are eligible for partial support when assembly and
annotation are publicly available through Genbank. The
assemblies, annotations, and functional data tracks (GFF files)
will be available on the JBrowse genome browser and can be
searched via BLAST.
The Echinobase PIs and SAB members will decide the new
species for full support and integration into the database as
gene pages with complete functionality, including gene names and
symbols, synonyms, literature, orthology, and developmental gene
expression display. This process will only be possible when the
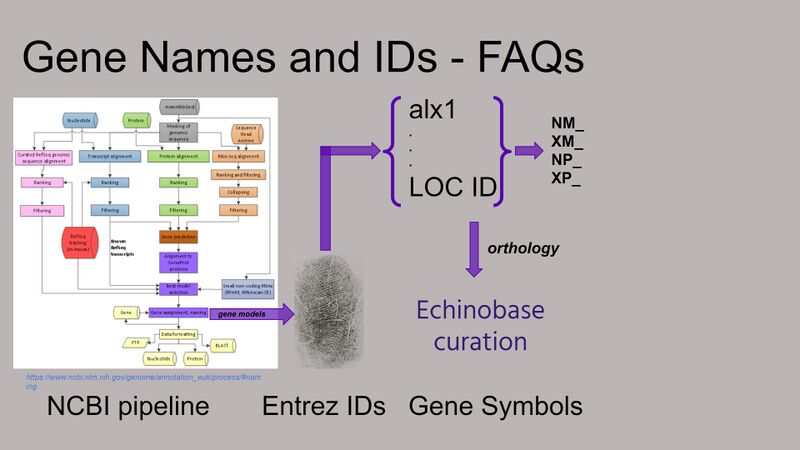
genome has been fully processed by the NCBI annotation pipeline.
Suggestions for priority species are always welcome.